In Charles Babbage’s Passages from the Life of a Philosopher he recalls two incidents where he is asked, “Pray, Mr. Babbage, if you put into the machine wrong figures, will the right answers come out?” Reflecting on these incidents he comments, “I am not able rightly to apprehend the kind of confusion of ideas that could provoke such a question.” Similarly, accurate and clean data is at the core of a functional AI model. However, ensuring accuracy of input data to avoid any “wrong figures” creeping into training datasets used to train AI models, demands meticulous attention during the stages of data wrangling, cleaning, and curation.
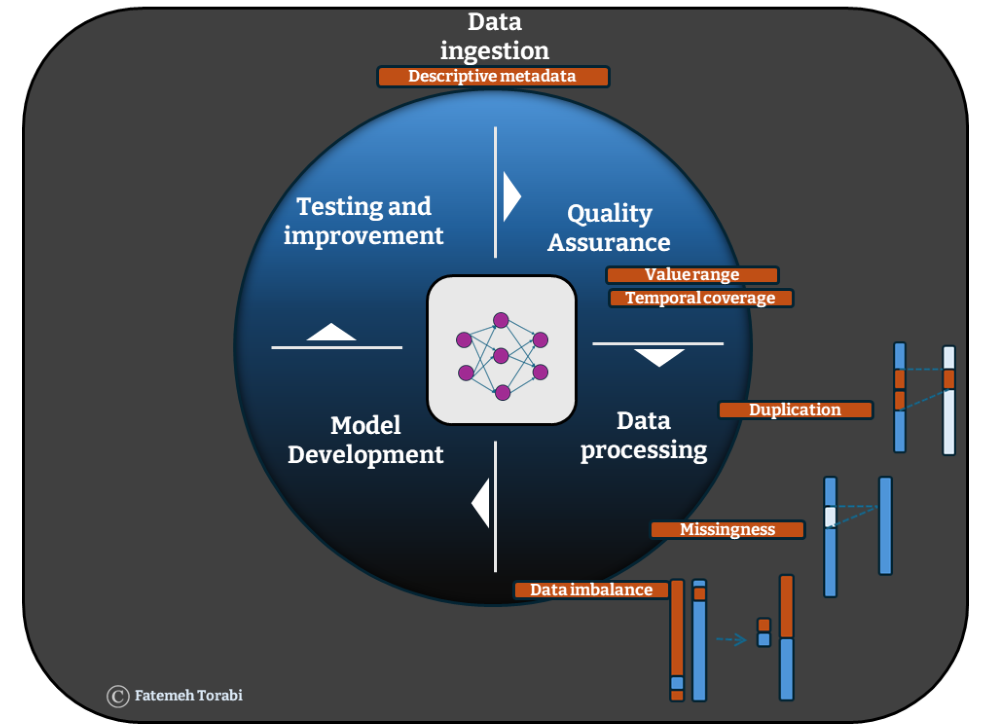
This necessity is particularly pronounced when dealing with the vast datasets used for training machine learning algorithms which are at the core of AI models. The predictive power of these models are highly dependent on the quality of the training data. 1 The most obvious errors which often require meticulous attention at data processing stages are duplication, missingness and data imbalance (Figure 1). The presence of any of these errors can exert multifaceted impacts both in the training and testing stage of machine learning algorithms that are at the core of any AI models, and the challenge does not end there. The provenance, content, format and structure of the data require attention as well. Even data that is essentially correct may be “wrong” for a particular data set.
Obviating the obvious errors
Duplicated records can mask existing diversities within the data, diminishing the representativeness of important subgroups and leading to a biased training set and model outcomes. If duplication originates from data labelling issues, it can lead to fundamental challenges during the training of supervised models. 2 In healthcare data, this issue can arise when linking data across multiple sources where each source holds different labels for the same data. 3
Missingness directly leads to loss of information required for training the algorithms on various real-world scenarios. It is typically addressed via two primary routes: deletion of missing rows or imputation. Deleting missing rows results in a reduction of the sample size and bias. For instance, when it comes to health data, using electronic medical records from a single health provider such as general practice may give rise to a lot of missingness in other aspects of an individual’s health such as their hospital records, pathology testing data or medical imaging. On the one hand, structured missingness can serve as an informative feature to explore within the data. However in cases where missing data causes pixelation in the comprehensive health picture we are attempting to construct, it often conceals an underlying narrative. 4 For instance, the COVID-19 response involved many initiatives across the AI community, however, during the early stages of the pandemic partial availability of data pixelated the picture and impacted models predictive ability which resulted in a minimal improvement according to the UK’s national centre for data science and AI report. 5
Imputing missing values can preserve the whole sample. However, the introduction of noise during the imputation process may compromise the quality of fitted models, contingent upon the proportion of missing records. One way to offset this may be to use larger and larger data sets that might inevitably include a fuller training picture to the algorithm, just as adding dots to a pixelated image makes it more and more clear what is depicted.
It is often perceived that for certain instances, particularly in the context of deep learning models, such as neural networks, the model itself is capable of handling missing values without explicit imputation or deletion procedures. 6 Is this really true in a real-world scenario? Are these models advanced enough to achieve an optimised performance even when data quality is not at an optimised level? 7 Köse et al. (2020) investigated the effect of two conventional imputation approaches: Multiple imputation (MICE) 8 and Factor analysis of mixed data (FAMD) 9 on performance of deep learning models. Their study endorsed the use of such explicit imputation approaches, showing an enhancement in model performance. 10
Data imbalance issues, arise within datasets where there is a disproportionate amount of information pertaining to a specific aspect. When imbalanced data, rich in focused information, is used to train an AI model, the model becomes adept at learning about the specific aspect but may struggle to generalise its findings to diverse scenarios, thus fostering overfitting where the model achieves accurate prediction on the training data but loses accuracy for any new test dataset.
Overfitting severely undermines the predictive performance of AI models on data beyond their training set, defeating the primary purpose of these models. For instance, out of all strokes that occur, approximately 85% are ischaemic, caused by blockage of blood supply to part of the brain, and 15% are haemorrhagic, caused by bleeding into the brain. Development of machine-learning and AI-based stroke predictive models are therefore being affected by this natural imbalance in the two types of stroke. 11 This type of imbalance also exists in population wide studies where stroke itself is only present in a minority subsection of a healthy population. 12
The circumstances of data collection can lead to bias, so care is needed at early stages to ensure that datasets are representative of the real world. These types of error can be picked up at an initial Quality Assurance (QA) stage conducted to reveal any unexpected errors in data used by AI models. QA checks often involve principle checks on presented values to ensure they are the right data type and within the expected range and have the expected temporal coverage.
Finally the choice of features included in a data set requires consideration since this can also have implications on an algorithm. Taking another example from the COVID-19 pandemic, a group of researchers trained an algorithm on radiological imaging of COVID-19 patients where the position of the patient during radiology was present as a feature in the dataset. However, since more severe cases were lying down and less severe cases were standing up, the existence of this feature resulted in an algorithm that predicted COVID-19 risk based on the position of the patients. Here although the data included was correct, its inclusion in the dataset proved to be “wrong”.
Things get complicated
When AI algorithms encounter complex, unstructured data, the task of quality assurance suddenly balloons beyond tackling the three main errors highlighted above. Such circumstances require some kind of quality enhancement procedure, where datasets in the form of images or unstructured text go through a curation process which involves enhancement of the quality and standardisation of the format to the level required for integration into AI algorithms. This process of standardization of data is paramount across various domains, especially in healthcare where complex, unstructured health data holds transformative potential for AI driven advances that revolutionise diagnosis, treatment, and prognosis of diseases. From electronic health records to magnetic resonance imaging (MRI) scans and genetic sequences, this data offers a wealth of insights for AI models to learn from. Adopting standardised formats not only facilitates seamless integration of diverse datasets but also streamlines the development and deployment of AI models. However, unlocking this potential requires a strong foundation in high-quality, processed data which begins with standardisation.
One category of complex health data is neuroimaging of which a prime example is MRI. Different institutions will often employ different acquisition protocols and different ways of collecting and storing neuroimaging data. Above all, this can make it very difficult to integrate into existing workflows and processing pipelines, but it also makes it challenging to understand, compare and combine with other datasets. To address these challenges, the neuroimaging community has adopted the Brain Imaging Data Structure (BIDS) 13 – a standardised format for organising and naming MRI data which allows compliant data to integrate smoothly with existing workflows and AI models, streamlining processing and analysis. By embracing standardisation, we can pave the way for common processing tools to enable the generation of AI-ready data.
Next, comes pre-processing. Sticking with the neuroimaging example, MRI scans are susceptible to various forms of noise and artifacts, which can appear as blurring or distortions which, without proper processing, can be misinterpreted by AI models. Pre-processing typically includes steps for spatial normalisation and image registration, involving alignment of brain images from different individuals into a common reference model and alignment of different images of the same subject to a common template. This standardisation facilitates inter-subject and inter-study comparisons, enabling AI models to generalise effectively across diverse datasets. However, the multi-layer aspect of this process means that aligning data to a common template is dependent on the choice of template which itself can introduce bias if the template brain doesn’t accurately reflect the patient’s anatomy (due to age, ethnicity, or disease for example).

Once pre-processing is complete, you may want to combine datasets to increase sample sizes for your AI model. This is where harmonisation techniques 14 15 come in to deal with inconsistencies and variations in acquisition which can add noise and bias into a model. A typical technique for harmonisation in neuroimaging, known as ComBat 16, works by modelling data using a hierarchical Bayesian model and followed by empirical Bayes to infer the distribution of the unknown factors. The method is actually borrowed from genomics data but is applicable to situations where multiple features of the same type are collected for each participant, whether that be expression levels for genes, or imaging derived measures such as volumes from different regions. This is a crucial step for combining datasets to enable AI models to focus on learning the actual relationships within the data rather than struggling with inconsistencies across datasets. It also leads to models which can generalise better on unseen data.
Feeding hungry algorithms
The public good is at the heart of AI driven approaches and indeed, the aim is to develop models with optimised predictive ability that can be generalised to many scenarios. For this to be achieved a large and diverse training source is required. This is often referred to as data hungry algorithms. To provide a large amount of enriched training data for optimised model development two main approaches have been explored: federated analytics and synthetic data.
Federation is when data from multiple sources is made available for training and analysis of models designed to run on data that is not held in a single place, nominally called distributed models. It provides the opportunity to test algorithms in different populations/settings to ensure generalisability. In the context of patient-level health data, the data is often held institutionally. Enabling federation and trustworthy sharing of these datasets requires extensive attention to governance models and a common model between multiple organisations is a known catalyst of this process 17 18
Generating synthetic data 19 from original data sources is a resource intensive mechanism. It requires the development of models on the real data to learn existing patterns, formats, and statistical properties within the original data from which it is possible to generate further synthetic versions of these data. When working with sensitive data such as health records, ensuring patient data is safe and secure is covered by information governance. Depending on how close the synthetic data source is to the original data, the same governance level may still be applicable when trying to bring individual/patient data from multiple sources together. A suggested solution to overcome the governance challenges in the context of synthetic data is to use a low-fidelity version of the original data which means a level of bias has been added throughout the synthesisation process to ensure safety and security of individual level data. 20 While the low fidelity data sources are generated based on real data, it is worth noting that the rise in generative AI also poses a concern for data pollution, particularly where AI tools such as Gretel.ai 21 are used to generate synthetic data which may also be used to train AI models – the problematic case of AI training AI!
When using sensitive health data of patients a further layer is in place to ensure security of access. These are called Trusted Research Environments (TREs), secure technology infrastructures which play a crucial role in consolidating disparate data collections into a centralised repository, facilitating researcher access to data for scientific exploration. However, integrating data from various sources into AI models poses challenges due to differences in data collection methods and formats, hindering computational analysis. In response, the FAIR (Findable, Accessible, Interoperable, Reusable) data principles were introduced in 2016 to enhance the reusability of scientific datasets by humans and machines. 22 Adoption of FAIR principles within TREs ensures well-documented, curated, and harmonised datasets, addressing issues raised above such as duplicated records and missing data. 23 Additionally, preprocessing pipelines within TREs streamline data standardisation, creating “AI research-ready” datasets. 24
Access to real-world healthcare data remains challenging, prompting the development of AI models on open-source or synthetic datasets. However, these models often exhibit performance discrepancies when applied to real world data 25 It is therefore imperative to provide researchers with secure access to real-world healthcare data within TREs, bolstered by robust governance and support mechanisms. Initiatives like the GRAIMATTER study 26 and AI risk evaluation workshops 27 exemplify efforts to facilitate AI model development and translation from TREs to clinical settings. By establishing governance guidelines and promoting FAIR datasets, TREs aim to become important resources for the AI research community. Providing standardised and curated data rich repositories that AI models can be developed on is a top priority in UK-TREs. Given the well-defined and secure governance environments of TREs they may also provide the basis for federated data analysis allowing researchers to combine datasets across TREs/data environments. In this way they can provide the large numbers that data hungry algorithms require, while avoiding the wide-ranging and myriad ways that data for a specific dataset can be “wrong”.
Also in the AI series:
What is AI? Shedding light on the method and madness in these algorithms Generative AI models and the quest for human-level artificial intelligence
- About the authors
- Fatemeh Torabi is Senior Research Officer and Data Scientist, at Swansea University and works on Health Data Science and Population Data Science for the Dementias Platform UK.
-
Lewis Hotchkiss is a Research Officer in Neuroimaging at Swansea University and works on Population Data Science for the Dementias Platform UK.
-
Emma Squires is the Data Project Manager for Dementias Platform UK based at Swansea University and works on Population Data Science
-
Prof. Simon E. Thompson is Deputy Associate Director of the Dementias Platform UK
-
Prof. Ronan A. Lyons is the Associate Director of the Dementias Platform UK based at Swansea University and works on Population Data Science.
- Copyright and licence
- © 2024 Royal Statistical Society
This article is licensed under a Creative Commons Attribution 4.0 (CC BY 4.0) International licence.
- How to cite
- Torabi, Fatemeh, Hotchkiss, Lewis, Squires, Emma, Thompson, Simon E. and Lyons, Ronan A. 2024. “Getting the data right for optimised AI performance.” Real World Data Science, May 7, 2024. URL
Footnotes
Li, P. et al. CleanML: A Benchmark for Joint Data Cleaning and Machine Learning [Experiments and Analysis]↩︎
Azeroual, O. et al. A Record Linkage-Based Data Deduplication Framework with DataCleaner Extension. Multimodal Technol. Interact. 2022, Vol. 6, Page 27 6, 27 (2022).↩︎
Rajpurkar, P., Chen, E., Banerjee, O. & Topol, E. J. AI in health and medicine. Nat. Med. 2022 281 28, 31–38 (2022).↩︎
Mitra, R. et al. Learning from data with structured missingness. (2023).↩︎
Alan Turing Institution. Data science and AI in the age of COVID-19. 2020 https://www.turing.ac.uk/sites/default/files/2021-06/data-science-and-ai-in-the-age-of-covid_full-report_2.pdf↩︎
Han, J. & Kang, S. Dynamic imputation for improved training of neural network with missing values. Expert Syst. Appl. 194, 116508 (2022).↩︎
Köse, T. et al. Effect of Missing Data Imputation on Deep Learning Prediction Performance for Vesicoureteral Reflux and Recurrent Urinary Tract Infection Clinical Study. Biomed Res. Int. 2020, (2020).↩︎
Azur, M. J., Stuart, E. A., Frangakis, C. & Leaf, P. J. Multiple imputation by chained equations: what is it and how does it work? Int. J. Methods Psychiatr. Res. 20, 40–49 (2011).↩︎
Audigier, V., Husson, F. & Josse, J. A principal component method to impute missing values for mixed data. Adv. Data Anal. Classif. 10, 5–26 (2016).↩︎
Köse, T. et al. Effect of Missing Data Imputation on Deep Learning Prediction Performance for Vesicoureteral Reflux and Recurrent Urinary Tract Infection Clinical Study. Biomed Res. Int. 2020, (2020).↩︎
Liu, T., Fan, W. & Wu, C. A hybrid machine learning approach to cerebral stroke prediction based on imbalanced medical dataset. Artif. Intell. Med. 101, 101723 (2019).↩︎
Kokkotis, C. et al. An Explainable Machine Learning Pipeline for Stroke Prediction on Imbalanced Data. Diagnostics 2022, Vol. 12, Page 2392 12, 2392 (2022).↩︎
Gorgolewski, K. J. et al. BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput. Biol. 13, (2017).↩︎
Bauermeister, S. et al. Research-ready data: the C-Surv data model. Eur. J. Epidemiol. 38, 179–187 (2023).↩︎
Abbasizanjani, H. et al. Harmonising electronic health records for reproducible research: challenges, solutions and recommendations from a UK-wide COVID-19 research collaboration. BMC Med. Inform. Decis. Mak. 23, 1–15 (2023).↩︎
Orlhac, F. et al. A Guide to ComBat Harmonization of Imaging Biomarkers in Multicenter Studies. J. Nucl. Med. 63, 172 (2022).↩︎
Toga, A. W. et al. The pursuit of approaches to federate data to accelerate Alzheimer’s disease and related dementia research: GAAIN, DPUK, and ADDI. Front. Neuroinform. 17, 1175689 (2023).↩︎
Torabi, F. et al. A common framework for health data governance standards. Nat. Med. 2024 1–4 (2024) doi:10.1038/s41591-023-02686-w.↩︎
Tucker, A., Wang, Z., Rotalinti, Y. & Myles, P. Generating high-fidelity synthetic patient data for assessing machine learning healthcare software. npj Digit. Med. 2020 31 3, 1–13 (2020)↩︎
Tucker, A., Wang, Z., Rotalinti, Y. & Myles, P. Generating high-fidelity synthetic patient data for assessing machine learning healthcare software. npj Digit. Med. 2020 31 3, 1–13 (2020)↩︎
Noruzman, A. H., Ghani, N. A. & Zulkifli, N. S. A. Gretel.ai: Open-Source Artificial Intelligence Tool To Generate New Synthetic Data. MALAYSIAN J. Innov. Eng. Appl. Soc. Sci. 1, 15–22 (2021).↩︎
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016 31 3, 1–9 (2016).↩︎
Chen, Y. et al. A FAIR and AI-ready Higgs boson decay dataset. Sci. Data 9, (2021).↩︎
Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2018).↩︎
Alkhalifah, T., Wang, H. & Ovcharenko, O. MLReal: Bridging the gap between training on synthetic data and real data applications in machine learning. Artif. Intell. Geosci. 3, 101–114 (2022).↩︎
Jefferson, E. et al. GRAIMATTER Green Paper: Recommendations for disclosure control of trained Machine Learning (ML) models from Trusted Research Environments (TREs). doi:10.5281/ZENODO.7089491.↩︎
DARE UK Community Working Group - DARE UK. https://dareuk.org.uk/dare-uk-community-working-groups/dare-uk-community-working-group-ai-risk-evaluation-working-group/.↩︎